Using PyMC
PyMC is a very powerful Python library designed for probabilistic and Bayesian analysis.
Here, we show that PyMC can be used to perform the same likelihood sampling that we previously wrote our own algorithm for.
Below, we read in the data and build the model.
The next step is to construct the PyMC sampler.
The format that PyMC expects can be a bit unfamiliar.
First we create objects for the two parameters, these are bounded so \(0 \leq k < 1\) and \(0 \leq [A]_0 < 10\) .
Strictly, these are prior probabilities tune
Unlike the code that we created previously, PyMC defaults to using the NUTS sampler, which stands for No-U-Turn sampler [7 ] .
This sampler enables the step size tuning that we have taken advantage of.
This results in a object assigned to the variable trace
posterior
<xarray.Dataset> Size: 168kB
Dimensions: (chain: 10, draw: 1000)
Coordinates:
* chain (chain) int64 80B 0 1 2 3 4 5 6 7 8 9
* draw (draw) int64 8kB 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999
Data variables:
k (chain, draw) float64 80kB 0.09838 0.0997 0.1063 ... 0.1197 0.09881
A0 (chain, draw) float64 80kB 7.332 7.07 7.475 ... 7.88 7.88 6.942
Attributes:
created_at: 2025-08-21T09:19:10.700373+00:00
arviz_version: 0.22.0
inference_library: pymc
inference_library_version: 5.20.0
sampling_time: 5.05695366859436
tuning_steps: 1000 Dimensions:
Coordinates: (2)
Data variables: (2)
Indexes: (2)
PandasIndex
PandasIndex(Index([0, 1, 2, 3, 4, 5, 6, 7, 8, 9], dtype='int64', name='chain')) PandasIndex
PandasIndex(Index([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9,
...
990, 991, 992, 993, 994, 995, 996, 997, 998, 999],
dtype='int64', name='draw', length=1000)) Attributes: (6)
created_at : 2025-08-21T09:19:10.700373+00:00 arviz_version : 0.22.0 inference_library : pymc inference_library_version : 5.20.0 sampling_time : 5.05695366859436 tuning_steps : 1000
sample_stats
<xarray.Dataset> Size: 1MB
Dimensions: (chain: 10, draw: 1000)
Coordinates:
* chain (chain) int64 80B 0 1 2 3 4 5 6 7 8 9
* draw (draw) int64 8kB 0 1 2 3 4 5 ... 995 996 997 998 999
Data variables: (12/17)
largest_eigval (chain, draw) float64 80kB nan nan nan ... nan nan
step_size (chain, draw) float64 80kB 0.8339 0.8339 ... 0.5714
perf_counter_start (chain, draw) float64 80kB 1.101e+03 ... 1.105e+03
energy_error (chain, draw) float64 80kB 0.03355 ... 0.04462
smallest_eigval (chain, draw) float64 80kB nan nan nan ... nan nan
max_energy_error (chain, draw) float64 80kB 0.09537 0.2051 ... -0.4004
... ...
diverging (chain, draw) bool 10kB False False ... False False
n_steps (chain, draw) float64 80kB 3.0 3.0 5.0 ... 1.0 5.0
energy (chain, draw) float64 80kB 3.633 4.11 ... 5.347 4.948
process_time_diff (chain, draw) float64 80kB 0.0003405 ... 0.0003039
acceptance_rate (chain, draw) float64 80kB 0.9373 0.8784 ... 0.956
index_in_trajectory (chain, draw) int64 80kB 2 2 1 2 -3 2 ... 2 -1 -1 0 3
Attributes:
created_at: 2025-08-21T09:19:10.721943+00:00
arviz_version: 0.22.0
inference_library: pymc
inference_library_version: 5.20.0
sampling_time: 5.05695366859436
tuning_steps: 1000 Dimensions:
Coordinates: (2)
Data variables: (17)
largest_eigval
(chain, draw)
float64
nan nan nan nan ... nan nan nan nan
array([[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]]) step_size
(chain, draw)
float64
0.8339 0.8339 ... 0.5714 0.5714
array([[0.83393177, 0.83393177, 0.83393177, ..., 0.83393177, 0.83393177,
0.83393177],
[0.50826497, 0.50826497, 0.50826497, ..., 0.50826497, 0.50826497,
0.50826497],
[0.64100498, 0.64100498, 0.64100498, ..., 0.64100498, 0.64100498,
0.64100498],
...,
[0.62172778, 0.62172778, 0.62172778, ..., 0.62172778, 0.62172778,
0.62172778],
[0.42439063, 0.42439063, 0.42439063, ..., 0.42439063, 0.42439063,
0.42439063],
[0.57140697, 0.57140697, 0.57140697, ..., 0.57140697, 0.57140697,
0.57140697]]) perf_counter_start
(chain, draw)
float64
1.101e+03 1.101e+03 ... 1.105e+03
array([[1100.88162556, 1100.88207987, 1100.8825198 , ..., 1101.33499953,
1101.3355021 , 1101.33598805],
[1100.68236968, 1100.68283376, 1100.6831518 , ..., 1101.05127719,
1101.05154605, 1101.05181964],
[1101.56621095, 1101.56669268, 1101.56697916, ..., 1102.02557072,
1102.02597439, 1102.02640152],
...,
[1103.70326081, 1103.70355608, 1103.70384476, ..., 1104.13214274,
1104.13241958, 1104.13269952],
[1104.42606019, 1104.42646632, 1104.42683149, ..., 1104.85463088,
1104.85514085, 1104.85575846],
[1104.59187249, 1104.5925352 , 1104.59298362, ..., 1104.96743087,
1104.96762908, 1104.96780438]]) energy_error
(chain, draw)
float64
0.03355 0.07476 ... 0.0 0.04462
array([[ 3.35547299e-02, 7.47641534e-02, -8.54076058e-02, ...,
-6.06665197e-02, -3.98526241e-02, 1.54061934e-01],
[ 5.29850800e-03, -4.75258488e-02, 1.29317746e-01, ...,
1.77996979e-01, -2.41800567e-01, 1.06466469e-01],
[-8.99009956e-02, 0.00000000e+00, 2.79109030e-01, ...,
-6.57230828e-01, -2.42393489e-04, 9.96117545e-01],
...,
[ 0.00000000e+00, -4.52493632e-01, 3.36870124e-01, ...,
-9.23319582e-01, -2.51420861e-01, 2.84582213e-02],
[ 1.71884234e-01, -1.99856560e-01, 1.14895123e-01, ...,
-3.71753838e-01, -8.72319373e-02, -1.86581539e-01],
[ 4.00792250e-01, 0.00000000e+00, -3.52711603e-01, ...,
-3.81319682e-01, 0.00000000e+00, 4.46247536e-02]]) smallest_eigval
(chain, draw)
float64
nan nan nan nan ... nan nan nan nan
array([[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]]) max_energy_error
(chain, draw)
float64
0.09537 0.2051 ... 0.6608 -0.4004
array([[ 9.53717221e-02, 2.05132056e-01, 8.77882288e-02, ...,
1.70308143e-01, 4.97757008e-02, 1.54061934e-01],
[-4.50009345e-02, 1.05615976e-01, 2.54348723e-01, ...,
1.83139006e-01, 3.27522346e-01, 2.74976892e-01],
[-1.14062418e-01, 7.47183000e-01, 2.79109030e-01, ...,
-7.00477410e-01, 1.09379342e-03, 1.06166205e+00],
...,
[ 2.54249470e+00, -4.52493632e-01, 6.15774611e-01, ...,
1.00930040e+00, -2.90219342e-01, -1.32350634e-01],
[ 3.54796938e-01, -1.99856560e-01, 1.31597228e-01, ...,
6.45933120e-01, 3.26647881e-01, -1.96597553e-01],
[ 4.00792250e-01, 6.30327456e+00, -3.63497454e-01, ...,
-3.81319682e-01, 6.60779632e-01, -4.00389067e-01]]) reached_max_treedepth
(chain, draw)
bool
False False False ... False False
array([[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]]) tree_depth
(chain, draw)
int64
2 2 3 2 2 3 2 2 ... 3 2 2 2 3 1 1 3
array([[2, 2, 3, ..., 2, 2, 2],
[3, 2, 3, ..., 2, 2, 2],
[3, 2, 3, ..., 2, 2, 2],
...,
[1, 1, 3, ..., 2, 2, 3],
[2, 2, 3, ..., 2, 3, 2],
[3, 2, 3, ..., 1, 1, 3]]) lp
(chain, draw)
float64
-3.492 -3.82 ... -4.412 -4.275
array([[-3.49249966, -3.8197008 , -3.29115402, ..., -4.21552395,
-4.41013982, -5.72781045],
[-3.35249495, -3.75337053, -4.71840053, ..., -7.16896418,
-5.83590392, -6.17853375],
[-3.30756475, -3.30756475, -4.58351028, ..., -3.24870943,
-3.25865278, -5.3328024 ],
...,
[-5.77327634, -4.92264088, -7.38567618, ..., -4.9329266 ,
-4.46492152, -4.58235085],
[-4.12244452, -3.43403727, -3.66427312, ..., -3.91016806,
-4.00027229, -3.90553002],
[-7.52177449, -7.52177449, -3.7156601 , ..., -4.41213165,
-4.41213165, -4.27544092]]) step_size_bar
(chain, draw)
float64
0.5881 0.5881 ... 0.5478 0.5478
array([[0.58809662, 0.58809662, 0.58809662, ..., 0.58809662, 0.58809662,
0.58809662],
[0.59462526, 0.59462526, 0.59462526, ..., 0.59462526, 0.59462526,
0.59462526],
[0.68055239, 0.68055239, 0.68055239, ..., 0.68055239, 0.68055239,
0.68055239],
...,
[0.66700707, 0.66700707, 0.66700707, ..., 0.66700707, 0.66700707,
0.66700707],
[0.68052707, 0.68052707, 0.68052707, ..., 0.68052707, 0.68052707,
0.68052707],
[0.5477832 , 0.5477832 , 0.5477832 , ..., 0.5477832 , 0.5477832 ,
0.5477832 ]]) perf_counter_diff
(chain, draw)
float64
0.0003402 0.0003306 ... 0.0003036
array([[0.00034016, 0.00033057, 0.0004593 , ..., 0.00040706, 0.00039399,
0.00029315],
[0.00038827, 0.00023705, 0.0003654 , ..., 0.00019525, 0.00020011,
0.00022371],
[0.00039719, 0.00020797, 0.00040359, ..., 0.00031306, 0.0003363 ,
0.00031341],
...,
[0.00019572, 0.00018056, 0.00062367, ..., 0.00020206, 0.00020445,
0.00042823],
[0.00030994, 0.00027267, 0.0005464 , ..., 0.00041665, 0.00050541,
0.00033838],
[0.00056793, 0.00034324, 0.00056324, ..., 0.00012535, 0.0001047 ,
0.00030364]]) diverging
(chain, draw)
bool
False False False ... False False
array([[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]]) n_steps
(chain, draw)
float64
3.0 3.0 5.0 3.0 ... 5.0 1.0 1.0 5.0
array([[3., 3., 5., ..., 3., 3., 3.],
[7., 3., 7., ..., 3., 3., 3.],
[7., 3., 7., ..., 3., 3., 3.],
...,
[1., 1., 7., ..., 3., 3., 7.],
[3., 3., 7., ..., 3., 7., 3.],
[7., 3., 7., ..., 1., 1., 5.]]) energy
(chain, draw)
float64
3.633 4.11 5.137 ... 5.347 4.948
array([[3.63253786, 4.11003318, 5.13673229, ..., 4.76159738, 5.10929628,
6.33739588],
[3.68011757, 4.70101482, 7.15745886, ..., 7.35519936, 8.16547504,
6.92693019],
[4.9644174 , 4.15681711, 4.60911394, ..., 4.37160325, 3.27528707,
5.43815692],
...,
[9.58578509, 6.14912896, 9.15282782, ..., 9.39738262, 5.48555532,
5.07297003],
[4.36850323, 4.26889649, 3.86653643, ..., 5.66330762, 5.46594772,
4.05444587],
[7.62344092, 8.72643502, 7.48434265, ..., 5.01449657, 5.34725399,
4.94809498]]) process_time_diff
(chain, draw)
float64
0.0003405 0.0003309 ... 0.0003039
array([[0.0003405 , 0.00033087, 0.0004596 , ..., 0.0003918 , 0.00039416,
0.00029343],
[0.00038834, 0.00023732, 0.00036569, ..., 0.00019548, 0.00020038,
0.00022387],
[0.00039725, 0.00020821, 0.00040391, ..., 0.00031333, 0.00033652,
0.00031366],
...,
[0.00019579, 0.00018084, 0.00062412, ..., 0.00020228, 0.00020474,
0.00042871],
[0.0003102 , 0.00027289, 0.00054658, ..., 0.00041671, 0.00050598,
0.00033828],
[0.00056796, 0.00034364, 0.00056383, ..., 0.00012485, 0.00010474,
0.00030388]]) acceptance_rate
(chain, draw)
float64
0.9373 0.8784 ... 0.5164 0.956
array([[0.93729791, 0.87842899, 0.98319097, ..., 0.93263028, 0.98381427,
0.91888179],
[0.99550181, 0.9536694 , 0.84135454, ..., 0.8850574 , 0.9069024 ,
0.85433309],
[1. , 0.65278433, 0.89558601, ..., 0.97872919, 0.99935709,
0.57173032],
...,
[0.0786699 , 1. , 0.65324092, ..., 0.74527296, 0.96205664,
0.99599184],
[0.75221242, 1. , 0.91912819, ..., 0.7399615 , 0.84371983,
0.9978955 ],
[0.86378243, 0.02079614, 1. , ..., 1. , 0.51644854,
0.95601569]]) index_in_trajectory
(chain, draw)
int64
2 2 1 2 -3 2 1 ... 1 3 2 -1 -1 0 3
array([[ 2, 2, 1, ..., -3, -3, -1],
[-4, -1, -2, ..., 2, 3, -2],
[-5, 0, 3, ..., 1, -1, 1],
...,
[ 0, 1, 3, ..., -2, -3, -3],
[-2, -1, 3, ..., 2, -2, -1],
[-3, 0, -3, ..., -1, 0, 3]]) Indexes: (2)
PandasIndex
PandasIndex(Index([0, 1, 2, 3, 4, 5, 6, 7, 8, 9], dtype='int64', name='chain')) PandasIndex
PandasIndex(Index([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9,
...
990, 991, 992, 993, 994, 995, 996, 997, 998, 999],
dtype='int64', name='draw', length=1000)) Attributes: (6)
created_at : 2025-08-21T09:19:10.721943+00:00 arviz_version : 0.22.0 inference_library : pymc inference_library_version : 5.20.0 sampling_time : 5.05695366859436 tuning_steps : 1000
observed_data
<xarray.Dataset> Size: 80B
Dimensions: (At_dim_0: 5)
Coordinates:
* At_dim_0 (At_dim_0) int64 40B 0 1 2 3 4
Data variables:
At (At_dim_0) float64 40B 6.23 3.76 2.6 1.85 1.27
Attributes:
created_at: 2025-08-21T09:19:10.726370+00:00
arviz_version: 0.22.0
inference_library: pymc
inference_library_version: 5.20.0
This contains the chain information amoung other things.
Instead of probing into the trace arviz
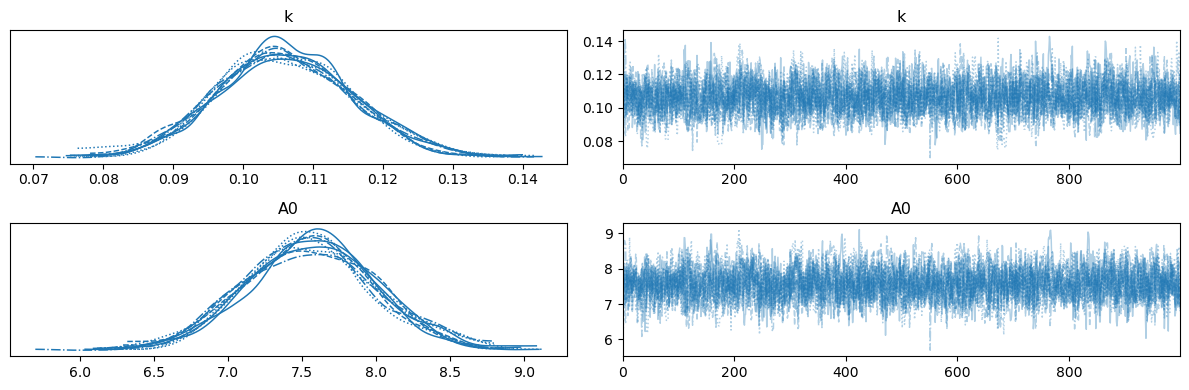
Above, we can see the trace of each of the different chains.
The chains appear to have converged to the same distribution.
We can get the flat chains with the following function.
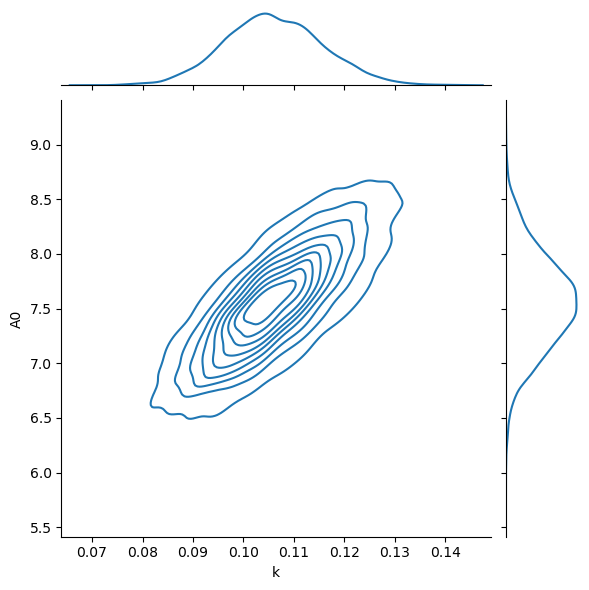
It is clear that, using PyMC, we have much better sampling of the distributions.
This makes using summary statistics, like the mean and standard deviation much more reliable.
mean
sd
hdi_3%
hdi_97%
mcse_mean
mcse_sd
ess_bulk
ess_tail
r_hat
k
0.106
0.01
0.088
0.124
0.000
0.000
2737.0
3531.0
1.0
A0
7.558
0.44
6.771
8.419
0.009
0.005
2678.0
3220.0
1.0