Ultranest for Nested Sampling#
We will look at the following data to investigate nested sampling using the ultranest package.
import pandas as pd
data = pd.read_csv('../data/signals.csv')
data.head()
| x | y | yerr | |
|---|---|---|---|
| 0 | 0 | -5.010978e-05 | 0.000079 |
| 1 | 4 | 1.952369e-05 | 0.000027 |
| 2 | 8 | 1.091299e-04 | 0.000130 |
| 3 | 12 | 3.387135e-09 | 0.000010 |
| 4 | 16 | -6.209843e-06 | 0.000051 |
This data is the measurement of some signal \(y\), as a function of \(x\).
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.errorbar(data['x'], data['y'], yerr=data['yerr'])
ax.set_xlabel('x')
ax.set_ylabel('y')
plt.show()
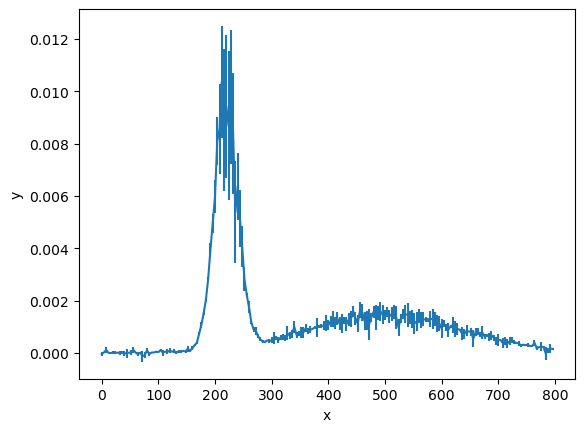
The question we want to answer about this data is if one or two Gaussian signals are present. We can think of this as two models: one for a single signal and a more complex one for two signals. This is the perfect opportunity to use Bayesian model selection.
Before constructing either model, we will create the likelihood function, which will be the same either way but with a different input model.
import numpy as np
from scipy.stats import norm
data_distribution = [norm(loc=loc, scale=scale) for loc, scale in zip(data['y'], data['yerr'])]
def likelihood(params, model):
"""
A general likelihood function for a model with Gaussian errors.
:param params: The parameters of the model.
:param model: The model function.
:return: The likelihood of the model given the data.
"""
model_y = model(data['x'], params)
return np.sum([d.logpdf(m) for d, m in zip(data_distribution, model_y)])
The next step is to build a simpler model and compute the evidence. We start by creating priors for the mean and standard deviation of the single Gaussian function.
from scipy.stats import uniform, norm
priors_one = [norm(220, 20),
uniform(10, 20)]
The mean has a normally distributed prior (\(\mathcal{N}(220, 20)\)) and the standard deviation is uniform (\(\mathcal{U}(10, 20)\)).
The next step is to create a prior transform function. This function converts uniformly distributed random variables to random variables drawn from the priors of interest.
def prior_transform_one(u):
"""
Transform the uniform random variables `u` to the model parameters.
:param u: Uniform random variables
:return: Model parameters
"""
return [p.ppf(u_) for p, u_ in zip(priors_one, u)]
Now, we construct the model and create a model-specific likelihood and function to be fed to the sampler.
def model_one(x, params):
"""
A simpler single Gaussian model.
:param x: The x values
:param params: The model parameters
:return: The y values
"""
return norm(loc=params[0], scale=params[1]).pdf(x)
def likelihood_one(params):
"""
The likelihood function for the simpler model.
:param params: The model parameters
:return: The likelihood
"""
return likelihood(params, model_one)
This is then passed to the ultranest.ReactiveNestedSampler.
import ultranest
sampler_one = ultranest.ReactiveNestedSampler(['mu1', 'sigma1'], likelihood_one, prior_transform_one)
sampler_one.run(show_status=False)
sampler_one.print_results()
[ultranest] Sampling 400 live points from prior ...
[ultranest] Explored until L=-7e+05
[ultranest] Likelihood function evaluations: 8208
[ultranest] logZ = -6.828e+05 +- 0.1103
[ultranest] Effective samples strategy satisfied (ESS = 1562.4, need >400)
[ultranest] Posterior uncertainty strategy is satisfied (KL: 0.46+-0.08 nat, need <0.50 nat)
[ultranest] Evidency uncertainty strategy is satisfied (dlogz=0.11, need <0.5)
[ultranest] logZ error budget: single: 0.15 bs:0.11 tail:0.01 total:0.11 required:<0.50
[ultranest] done iterating.
logZ = -682752.620 +- 0.268
single instance: logZ = -682752.620 +- 0.152
bootstrapped : logZ = -682752.596 +- 0.268
tail : logZ = +- 0.010
insert order U test : converged: True correlation: inf iterations
mu1 : 220.715│ ▁▁▁▁▁▁▁▁▂▂▃▄▄▅▆▆▇▆▇▆▆▅▄▃▃▂▁▁▁▁▁▁▁▁▁▁▁ │221.391 221.034 +- 0.084
sigma1 : 16.081│ ▁▁▁▁▁▁▁▁▁▁▂▃▃▅▄▆▇▇▇▇▇▇▆▆▅▅▄▃▂▁▁▁▁▁▁▁▁ │16.492 16.298 +- 0.053
sampler_one.results['logz'], sampler_one.results['logzerr']
(-682752.6198030798, 0.2678834502750158)
We can now do the same process for a more complex two-Gaussian model. Note that we have four priors now instead of two.
priors_two = [norm(220, 20),
uniform(10, 20),
norm(500, 100),
uniform(100, 200)]
def prior_transform_two(u):
"""
Transform the uniform random variables `u` to the model parameters for the more complex model.
:param u: Uniform random variables
:return: Model parameters
"""
return [p.ppf(u_) for p, u_ in zip(priors_two, u)]
def model_two(x, params):
"""
A more complex double Gaussian model.
:param x: The x values
:param params: The model parameters
:return: The y values
"""
return (norm(loc=params[0], scale=params[1]).pdf(x) + norm(loc=params[2], scale=params[3]).pdf(x)) * 0.5
def likelihood_two(params):
"""
The likelihood function for the more complex model.
:param params: The model parameters
:return: The likelihood
"""
return likelihood(params, model_two)
sampler_two = ultranest.ReactiveNestedSampler(['mu1', 'sigma1', 'mu2', 'sigma2'], likelihood_two, prior_transform_two)
sampler_two.run(show_status=False)
sampler_two.print_results()
[ultranest] Sampling 400 live points from prior ...
[ultranest] Explored until L=2e+03
[ultranest] Likelihood function evaluations: 22692
[ultranest] logZ = 1564 +- 0.1425
[ultranest] Effective samples strategy satisfied (ESS = 2135.9, need >400)
[ultranest] Posterior uncertainty strategy is satisfied (KL: 0.47+-0.07 nat, need <0.50 nat)
[ultranest] Evidency uncertainty strategy is satisfied (dlogz=0.14, need <0.5)
[ultranest] logZ error budget: single: 0.22 bs:0.14 tail:0.01 total:0.14 required:<0.50
[ultranest] done iterating.
logZ = 1563.889 +- 0.310
single instance: logZ = 1563.889 +- 0.223
bootstrapped : logZ = 1563.912 +- 0.310
tail : logZ = +- 0.010
insert order U test : converged: True correlation: inf iterations
mu1 : 219.22│ ▁ ▁▁▁▁▁▁▁▁▂▂▃▄▅▅▇▆▇▇▆▅▄▃▂▂▂▁▁▁▁▁▁▁▁▁ │220.49 219.88 +- 0.14
sigma1 : 19.46 │ ▁▁▁▁▁▁▁▁▂▂▂▄▄▅▆▇▇▇▇▇▆▇▅▄▃▃▂▁▁▁▁▁▁▁▁ ▁ │20.38 19.91 +- 0.11
mu2 : 498.12│ ▁ ▁▁▁▁▁▁▁▂▃▃▄▅▆▇▇▇▇▆▅▄▄▃▂▂▁▁▁▁▁▁▁▁▁▁ │502.75 500.40 +- 0.50
sigma2 : 137.63│ ▁▁▁▁▁▁▁▁▁▁▂▂▃▃▄▅▆▇▆▇▇▆▅▄▃▂▂▁▁▁▁▁▁▁ ▁ │141.55 139.63 +- 0.42
sampler_two.results['logz'], sampler_two.results['logzerr']
(1563.889404300776, 0.31049196778106025)
We can now calculate the Bayes factor for these two models.
log_B = sampler_two.results['logz'] - sampler_one.results['logz']
log_B
684316.5092073806
There is decisive evidence for the more complex model. Therefore, we would be able to continue our analysis by considering the signal shown by the two Gaussians.
